library(reticulate)
use_python("/Users/chammika/miniconda3/envs/science-work/bin/python", required = TRUE)Pandas (Good)
What to expect in this chapter
Now, we are ready to handle the topic of missing data. I delayed this discussion not because it is complicated but because I wanted to give you time to get comfortable with dataframes. We will continue to use the data from dummy-class_1-of-2.xlsx for this section.
1 Missing data
We know from info() that ‘Test 1’ and ‘Test 2’ are missing some data.
df_class.info()<class 'pandas.core.frame.DataFrame'>
Index: 35 entries, A3028967J to A7667457P
Data columns (total 7 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 MATRIC_NO 35 non-null object
1 Name 35 non-null object
2 Major 35 non-null category
3 Gender 35 non-null category
4 Test 1 33 non-null float64
5 Test 2 32 non-null float64
6 Total 31 non-null float64
dtypes: category(2), float64(3), object(2)
memory usage: 2.0+ KB1.1 All missing data
First, let’s see if we can get the exact locations of the missing data. Let’s start with the method isna().
df_class.isna()| MATRIC_NO | Name | Major | Gender | Test 1 | Test 2 | Total | |
|---|---|---|---|---|---|---|---|
| MATRIC_NO | |||||||
| A3028967J | False | False | False | False | False | False | False |
| A1282849W | False | False | False | False | False | False | False |
| A5408925A | False | False | False | False | False | False | False |
| A6973859L | False | False | False | False | False | False | False |
| A5410124H | False | False | False | False | False | True | True |
| A9568373Q | False | False | False | False | False | False | False |
| A6824244G | False | False | False | False | False | False | False |
| A9194090U | False | False | False | False | False | False | False |
| A4828364M | False | False | False | False | False | False | False |
| A4607700C | False | False | False | False | False | False | False |
| A7067766E | False | False | False | False | False | False | False |
| A5569996J | False | False | False | False | False | False | False |
| A3202548I | False | False | False | False | False | False | False |
| A6131593U | False | False | False | False | False | False | False |
| A7653832E | False | False | False | False | True | False | True |
| A9462811I | False | False | False | False | False | False | False |
| A1218599T | False | False | False | False | False | False | False |
| A7210476B | False | False | False | False | False | False | False |
| A1512479K | False | False | False | False | False | False | False |
| A7986368Y | False | False | False | False | False | False | False |
| A2727061A | False | False | False | False | False | True | True |
| A2999472W | False | False | False | False | False | False | False |
| A7116486E | False | False | False | False | False | False | False |
| A6931452S | False | False | False | False | False | False | False |
| A9649096H | False | False | False | False | False | False | False |
| A1643380L | False | False | False | False | False | False | False |
| A6787293E | False | False | False | False | False | False | False |
| A5975988J | False | False | False | False | False | False | False |
| A3699958T | False | False | False | False | True | True | True |
| A1956366U | False | False | False | False | False | False | False |
| A1468689D | False | False | False | False | False | False | False |
| A3217320C | False | False | False | False | False | False | False |
| A6867791C | False | False | False | False | False | False | False |
| A4080490P | False | False | False | False | False | False | False |
| A7667457P | False | False | False | False | False | False | False |
The missing data is where you see True. I hope that you will agree that this information is overwhelming and utterly useless. So, we need to have a way to get more refined information.
1.2 Columns and rows of the missing data
We can exercise more finesse by combining isna() with any(). any() in this case just asks if there are any True values. The axis option will allow us to pick the rows or columns (Honestly, I can never remember which is which, so I just try 0 or 1 and see what I get).
df_class.isna().any(axis=0) # Are there any True in the columns?MATRIC_NO False
Name False
Major False
Gender False
Test 1 True
Test 2 True
Total True
dtype: booldf_class.isna().any(axis=1) # Are there any True in the rows?MATRIC_NO
A3028967J False
A1282849W False
A5408925A False
A6973859L False
A5410124H True
A9568373Q False
A6824244G False
A9194090U False
A4828364M False
A4607700C False
A7067766E False
A5569996J False
A3202548I False
A6131593U False
A7653832E True
A9462811I False
A1218599T False
A7210476B False
A1512479K False
A7986368Y False
A2727061A True
A2999472W False
A7116486E False
A6931452S False
A9649096H False
A1643380L False
A6787293E False
A5975988J False
A3699958T True
A1956366U False
A1468689D False
A3217320C False
A6867791C False
A4080490P False
A7667457P False
dtype: bool1.3 More details of the missing numbers
Do you see that we can use the last output as a mask to locate the rows that have missing numbers? Let me show you the following:
mask_for_nan = df_class.isna().any(axis=1)
df_class[mask_for_nan]| MATRIC_NO | Name | Major | Gender | Test 1 | Test 2 | Total | |
|---|---|---|---|---|---|---|---|
| MATRIC_NO | |||||||
| A5410124H | A5410124H | Kyla Young | Physics | Male | 15.306 | NaN | NaN |
| A7653832E | A7653832E | Quentin Kemp | Chemistry | Female | NaN | 15.72 | NaN |
| A2727061A | A2727061A | Malik Becker | Physics | Male | 12.858 | NaN | NaN |
| A3699958T | A3699958T | Norah Miles | Chemistry | Male | NaN | NaN | NaN |
1.4 Dealing with missing numbers?
How you handle missing numbers must depend on what type of data you are dealing with.
Here are a few things you can do:
Replacing the missing data with a constant value
For our dummy class, the numbers are missing because the students were missing from the tests. So, it is easy; we give them zero (the fourth option). The easiest way to do this is using fillna(). I will use a crazy value first so that you can see what is going on.
df_class.loc[:, ['Test 1', 'Test 2', 'Total']].fillna(99999, inplace=True)
df_class[mask_for_nan]| MATRIC_NO | Name | Major | Gender | Test 1 | Test 2 | Total | |
|---|---|---|---|---|---|---|---|
| MATRIC_NO | |||||||
| A5410124H | A5410124H | Kyla Young | Physics | Male | 15.306 | NaN | NaN |
| A7653832E | A7653832E | Quentin Kemp | Chemistry | Female | NaN | 15.72 | NaN |
| A2727061A | A2727061A | Malik Becker | Physics | Male | 12.858 | NaN | NaN |
| A3699958T | A3699958T | Norah Miles | Chemistry | Male | NaN | NaN | NaN |
Do you see it? Yes, it’s that easy.
Let me first replace the crazy number with 0 before I forget.
df_class.replace(99999, 0, inplace = True)
df_class[mask_for_nan]| MATRIC_NO | Name | Major | Gender | Test 1 | Test 2 | Total | |
|---|---|---|---|---|---|---|---|
| MATRIC_NO | |||||||
| A5410124H | A5410124H | Kyla Young | Physics | Male | 15.306 | NaN | NaN |
| A7653832E | A7653832E | Quentin Kemp | Chemistry | Female | NaN | 15.72 | NaN |
| A2727061A | A2727061A | Malik Becker | Physics | Male | 12.858 | NaN | NaN |
| A3699958T | A3699958T | Norah Miles | Chemistry | Male | NaN | NaN | NaN |
Yes, there is a problem with the ‘Total’ column, but we can deal with that later since we have to modify it anyway.
Expunging all columns that have missing data (Drastic!)
I am not going to do this right now, but here is one way you can:
mask_for_nan_columns = df_class.isna().any(axis=0)
columns_to_keep = df_class.columns[~mask_for_nan_columns].values
df_class = df_class[columns_to_keep]I inverted (using ~) the mask to pick those columns that are NaN free. Then I just updated df_class with only those columns.
This works too:
df_class.dropna(axis = 0, inplace=True)Expunging all rows that have missing data (Drastic!)
Again, I am not going to do this right now, but here is one way you can:
df_class = df_class[~mask_for_nan]Again, I just inverted my mask. And, yet again, the following works too:
df_class.dropna(axis = 1, inplace=True)Replace missing data with a representative value (like an average)
Yet, again, I am not going to do this right now, but here is one way you can:
test_mean = df_class.loc['Test 1'].mean()
df.loc['Test 1'].fillna(test_mean)1.5 Itterating over a dataframe
I was looking for a way to introduce iterating (looping) over a dataframe. Now is an excellent opportunity because we can use it to replace the missing data. But this is certainly not the best way because there are highly optimised weapons in the Panda’s arsenal like apply(), fillna() and so on. However, I (lazily) often find myself using loops because they are plain and simple to get started and understand.
You can loop through your dataframe using the method itterrows() as follows. It spits out the index and the row for every iteration.
for index, row in df_class.iterrows():
name = row['Name']
for column in ['Test 1', 'Test 2']:
if np.isnan(row[column]):
print(f'{index}: {name:<15} missing data for {column}')
df_class.loc[index, column] = 0If you run this, you should end up with the following:
A5410124H: Kyla Young missing data for Test 2
A7653832E: Quentin Kemp missing data for Test 1
A2727061A: Malik Becker missing data for Test 2
A3699958T: Norah Miles missing data for Test 1
A3699958T: Norah Miles missing data for Test 2You can extract the various columns from the row variable just as you do with a dataframe (loc and iloc works too). I have used isnan() from NumPy to check for NaN because you cannot use ==. I am also using the index to update the original value of the dataframe. There are cautionary tales (e.g. see Nice of Loops) about changing something while iterating over it. However, it is safe in this case.
2 Combining dataframes
Now I will show you how to combine two datasets into one dataframe. Done correctly, Pandas makes quick, efficient work of this1.
The file dummy-class-2-of-2.xlsx contains the data for ‘Test 3’. You should open this file using Excel, LibreOffice or Sheets to see what it looks like.
I am going to import this data and call it df_class_2. I will also rename the columns and set the index to MATRIC_NO. This last point is crucial when combining the two dataframes because the index is used to match the rows.
df_class_2 = pd.read_excel('dummy-class-2-of-2.xlsx')
#------------------------- Rename columns ------------------------#
new_column_info = {'Student No': 'MATRIC_NO',
'Test 3 (50%)': 'Test 3'}
df_class_2.rename(columns=new_column_info, inplace=True)
#--------------------- Set index to MATRIC_NO --------------------#
df_class_2.set_index('MATRIC_NO', inplace=True)
df_class_2.info()<class 'pandas.core.frame.DataFrame'>
Index: 31 entries, A3028967J to A4080490P
Data columns (total 1 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Test 3 31 non-null float64
dtypes: float64(1)
memory usage: 496.0+ bytesIf you look at the output of info(), you will realise that students are missing! It turns out the person who created this file only included the scores for the students who were present at Test 3. If you were going to work with spreadsheets, this would have given you a migraine. But with Pandas, it is going to be a breeze.
Let me combine (concatenate) the two dataframe using the pd.concat() method. I will specify axis=1 so that the dataframes are joined along the columns (not one on top of each other along rows).
df_combined = pd.concat([df_class, df_class_2], axis=1)2.1 The complete recipe
Let me show you the complete recipe for everything we have done in this part. I have reordered some actions (e.g. creation of the ‘Total’ column) and modified a few things (e.g. df_class_1 instead of df_class). I have also removed the excessive comments from the previous version. Everything should flow nicely and make sense to you.
# -----------------------------------------------------------------#
# First file #
# -----------------------------------------------------------------#
df_class_1 = pd.read_excel('dummy-class-1-of-2.xlsx', skiprows=1)
columns_to_keep = ['Student No', 'Name', 'Major', 'Gender',
'Test 1 (30%)', 'Test 2 (20%)']
df_class_1 = df_class_1[columns_to_keep]
new_column_info = {'Student No': 'MATRIC_NO',
'Test 1 (30%)': 'Test 1',
'Test 2 (20%)': 'Test 2'}
df_class_1.rename(columns=new_column_info, inplace=True)
df_class_1.set_index('MATRIC_NO', drop=False, inplace=True)
replace_info = {
'PHY': 'Physics',
'CHM': 'Chemistry',
'LS': 'Life Sciences',
'CBIO': 'Comp. Biology',
'F': 'Female',
'M': 'Male',
'NB': 'Non-binary'
}
df_class_1.replace(to_replace=replace_info, inplace=True)
def clean(text):
'''
Function to remove ' ' from column 'Test 2'.
To be applied using apply()
'''
try:
return text.replace("'", "")
except AttributeError:
# This will handle the NaN of the missing data
return text
df_class_1['Test 2'] = df_class_1['Test 2'].apply(clean)
new_type_info = {'Major': 'category',
'Gender': 'category',
'Test 2': 'float'}
df_class_1 = df_class_1.astype(new_type_info)
# -----------------------------------------------------------------#
# Second file #
# -----------------------------------------------------------------#
df_class_2 = pd.read_excel('dummy-class-2-of-2.xlsx')
new_column_info = {'Student No': 'MATRIC_NO',
'Test 3 (50%)': 'Test 3'}
df_class_2.rename(columns=new_column_info, inplace=True)
df_class_2.set_index('MATRIC_NO', inplace=True)
# ---------- Concatenate the two dataframes along columns ---------#
df_combined = pd.concat([df_class_1, df_class_2], axis=1)
# -----------------------------------------------------------------#
df_combined.loc[:, ["Test 1", "Test 2", "Test 3"]].fillna(0, inplace=True)
df_combined["Total"] = df_combined[["Test 1", "Test 2", "Test 3"]].sum(axis=1)
df_combined.sort_values(by='Total', inplace=True)
df_combined = df_combined.round(2)
df_combined.to_excel('finalised_scores.xlsx', index=False)
df_combined.head()| MATRIC_NO | Name | Major | Gender | Test 1 | Test 2 | Test 3 | Total | |
|---|---|---|---|---|---|---|---|---|
| MATRIC_NO | ||||||||
| A3699958T | A3699958T | Norah Miles | Chemistry | Male | NaN | NaN | NaN | 0.00 |
| A6867791C | A6867791C | Katie Ayers | Comp. Biology | Female | 16.53 | 10.28 | NaN | 26.81 |
| A5410124H | A5410124H | Kyla Young | Physics | Male | 15.31 | NaN | 17.5 | 32.81 |
| A7210476B | A7210476B | Sidney Wiggins | Physics | Non-binary | 19.59 | 14.68 | NaN | 34.27 |
| A7667457P | A7667457P | Carter Crane | Life Sciences | Male | 20.82 | 13.82 | NaN | 34.63 |
3 Plotting directly from Pandas
Pandas allow us to speak directly to Matplotlib from the dataframe. So, the last thing I want to show you is how to do this as it will save you a lot of time. Especially when you are in the exploratory phase of using data.
For the following, I will use the df_combined dataframe. I will also keep my infatuation with xkcd in check and use another style from Seaborn2.
plt.style.use("seaborn-v0_8-darkgrid")3.1 Barcharts
-
grp = df_combined.groupby(['Gender'])["Total"] grp.mean().plot(kind='barh'); plt.tight_layout() plt.show()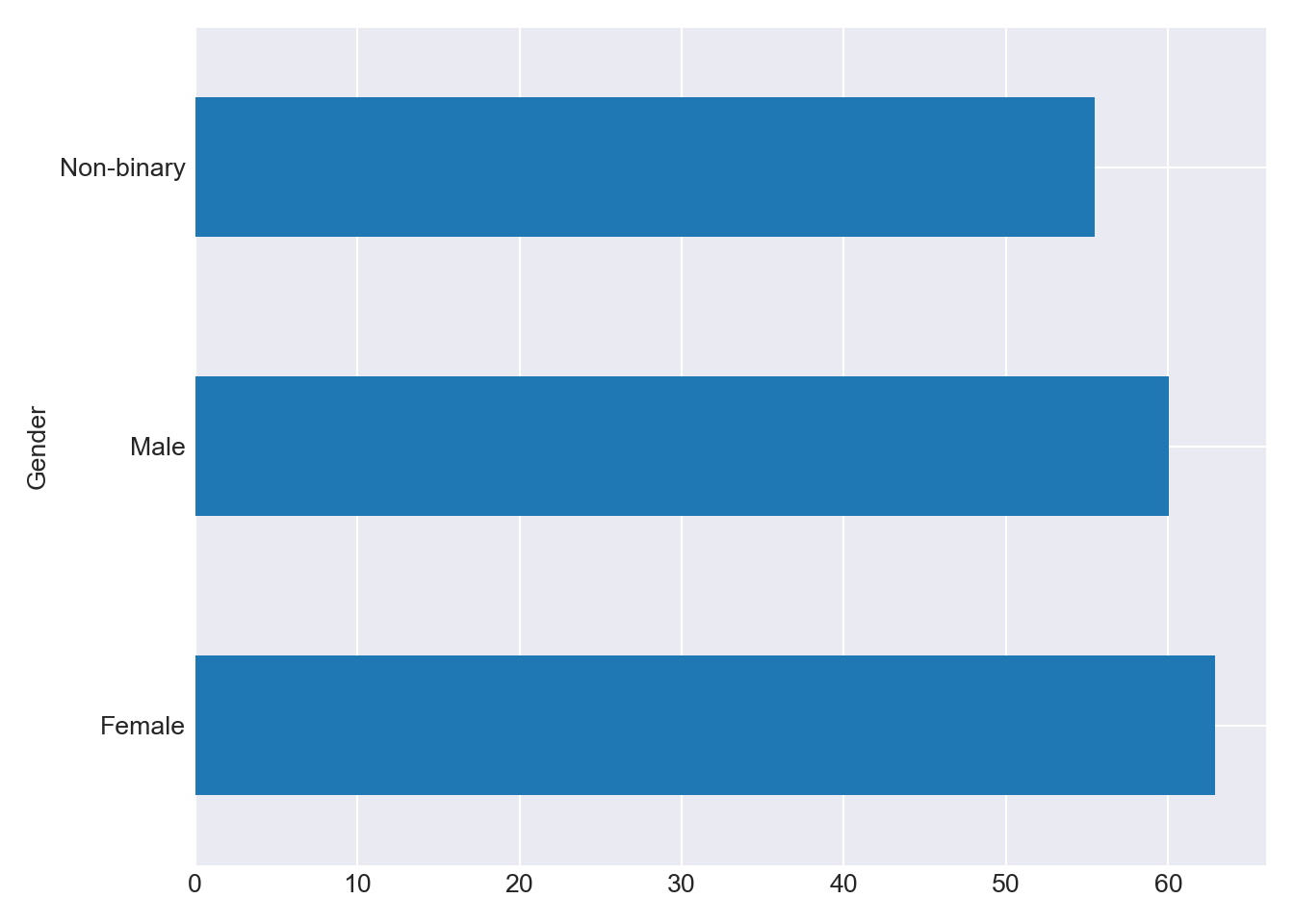
-
grp = df_combined.groupby(['Major'])[["Test 1", "Test 2", "Test 3"]] grp.mean().plot(kind='barh') plt.tight_layout() plt.show()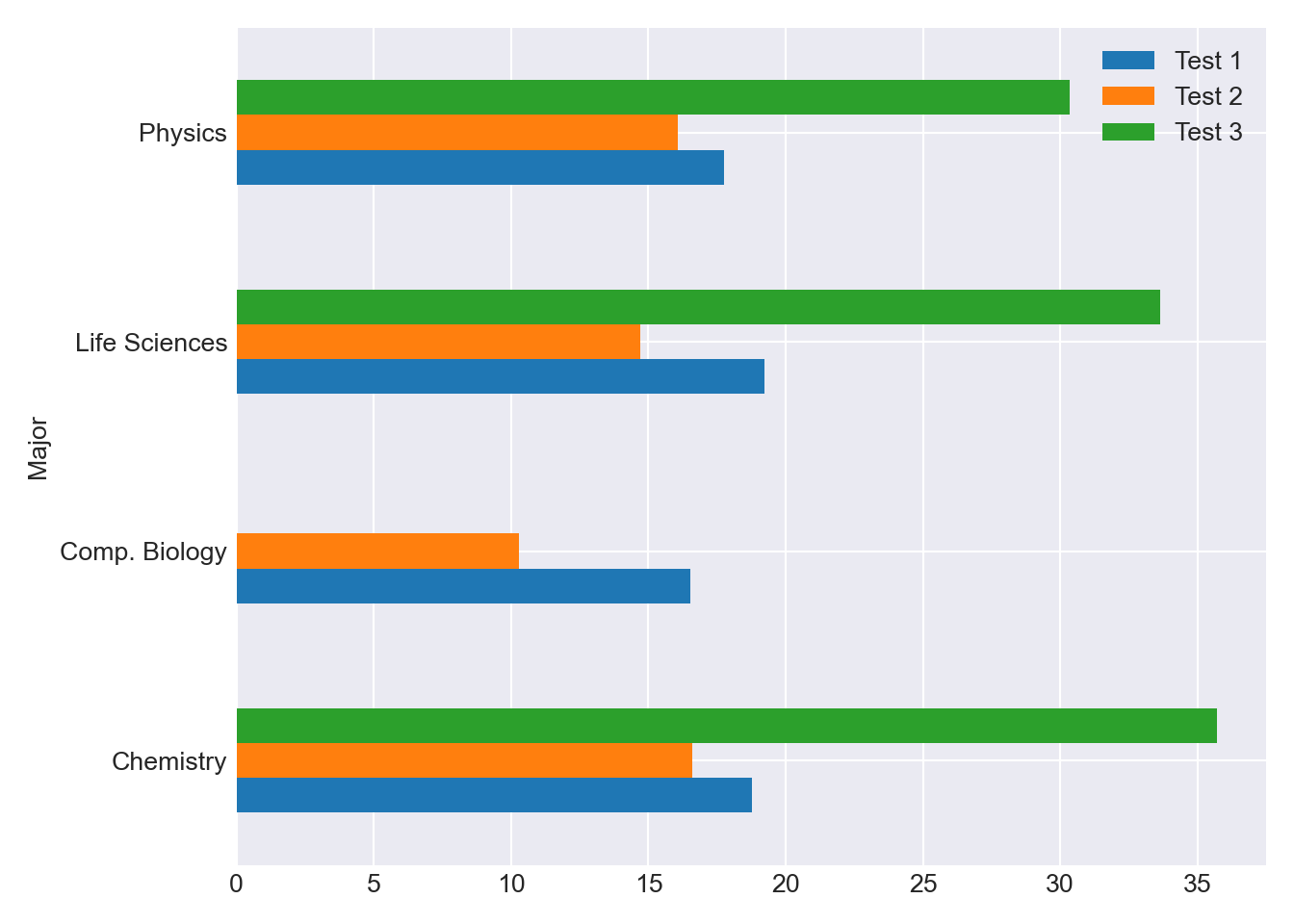
3.2 Histograms
-
If we like, we can create the axes that Pandas will use. Then, you can pass the axis using the
axoption. Let me show you.fix, ax = plt.subplots(nrows=2, ncols=2, sharey=True, figsize=(10,8)) df_combined['Total'].plot.hist(ax=ax[0, 0]) df_combined['Test 1'].plot.hist(ax=ax[0, 1]) df_combined['Test 2'].plot.hist(ax=ax[1, 0]) df_combined['Test 3'].plot.hist(ax=ax[1, 1]) plt.tight_layout() plt.show()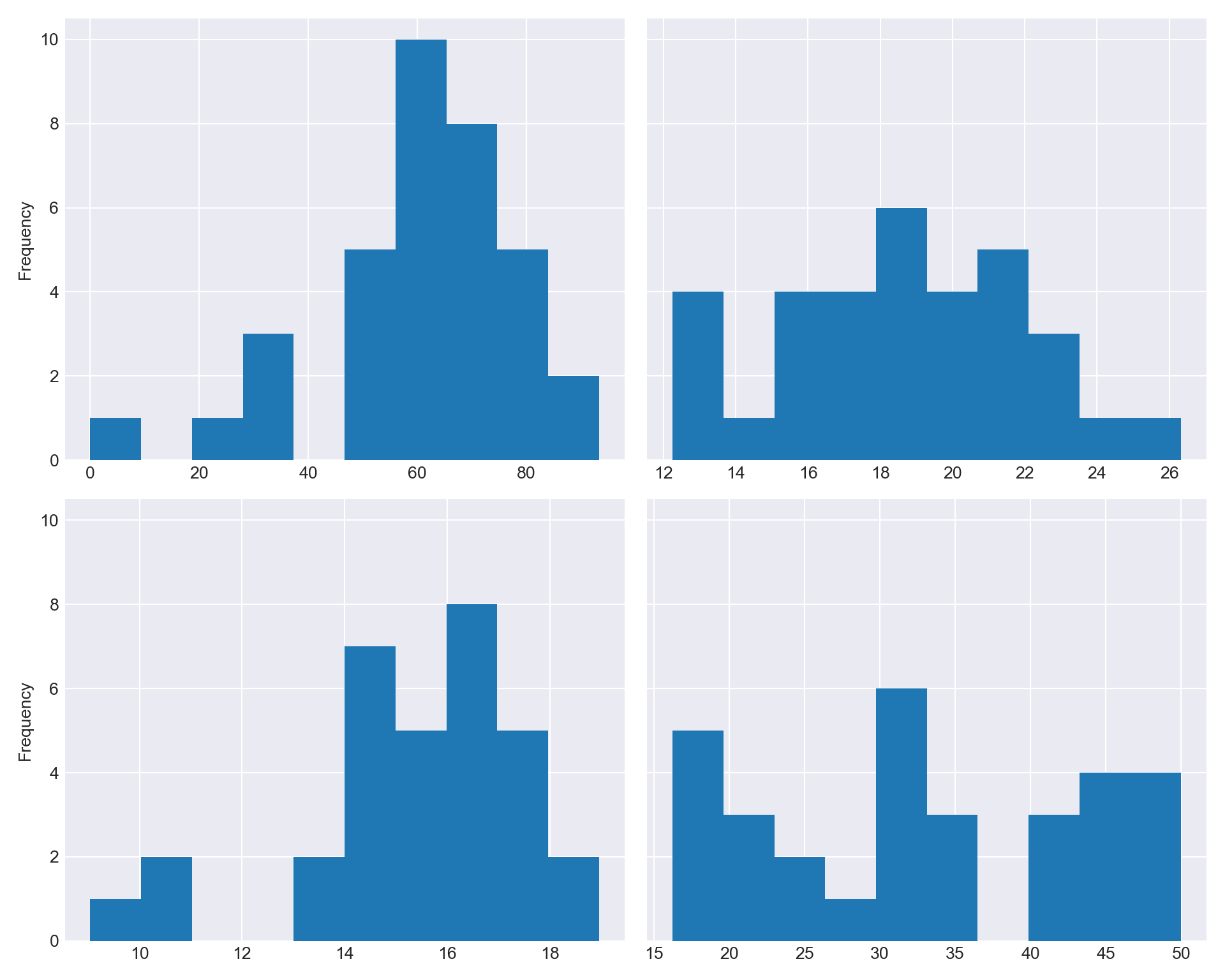
3.3 Pie
-
df_combined.groupby('Major').size().plot.pie(autopct="%d")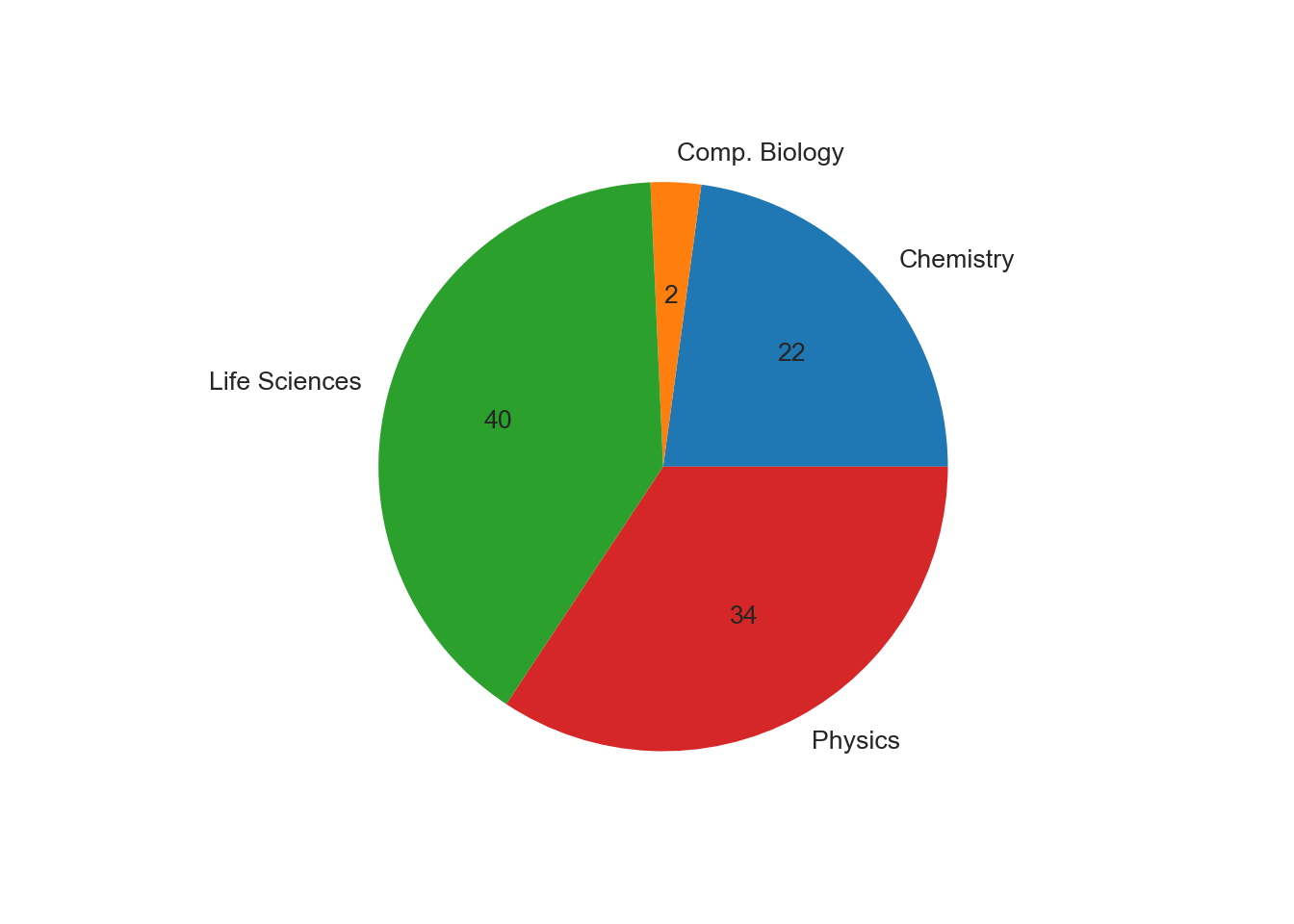
autopctspecifies the format in which the percentages are displayed.
3.4 Boxplots
Boxplots are amazing at conveying a lot of statistical information efficiently. You will not go wrong if you get into the habit of using them often.
-
This plots all the numerical columns as boxplots.
df_combined.boxplot() plt.show()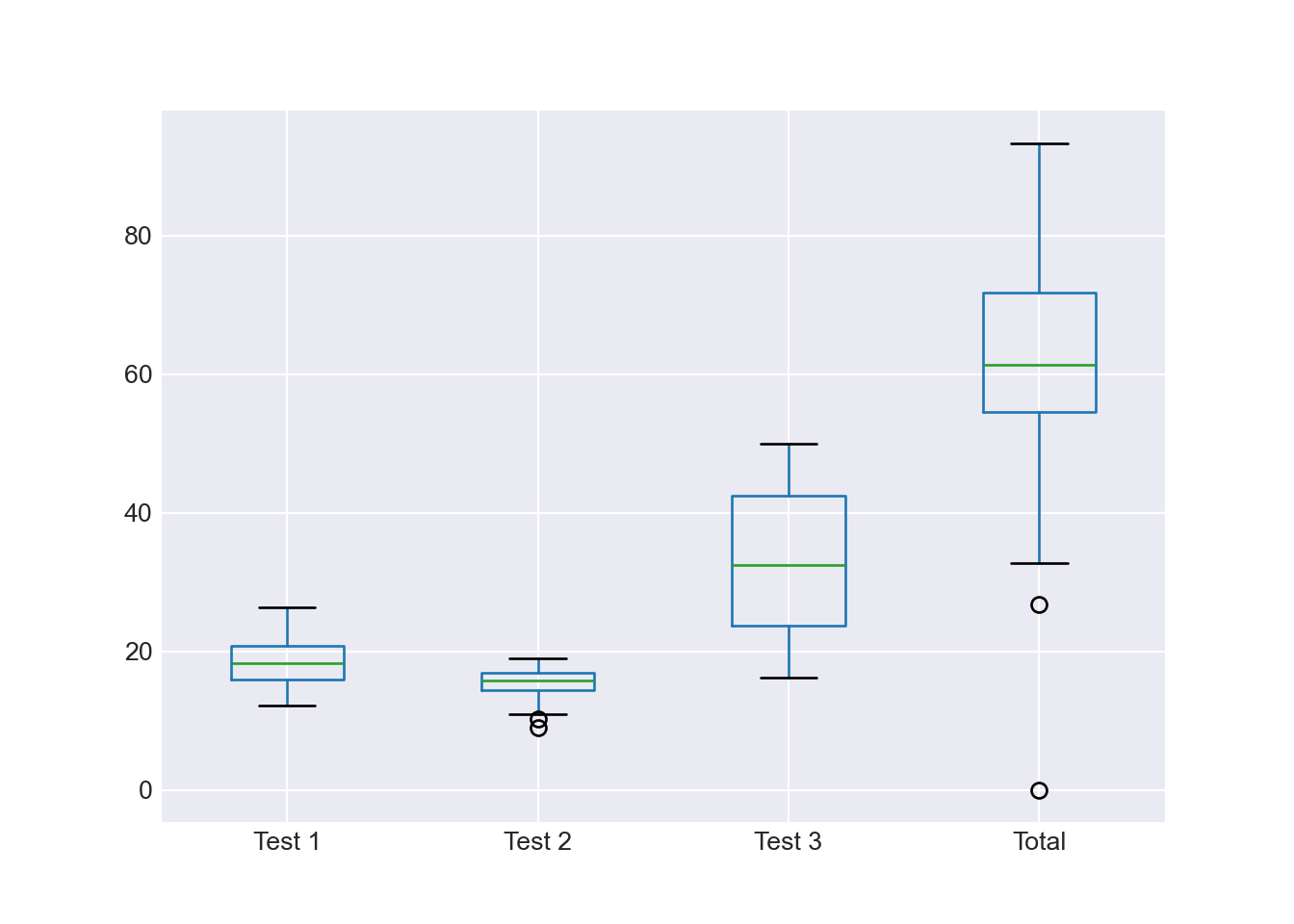
-
df_combined.boxplot(by='Major', column=['Total'], vert=False); plt.tight_layout() plt.show()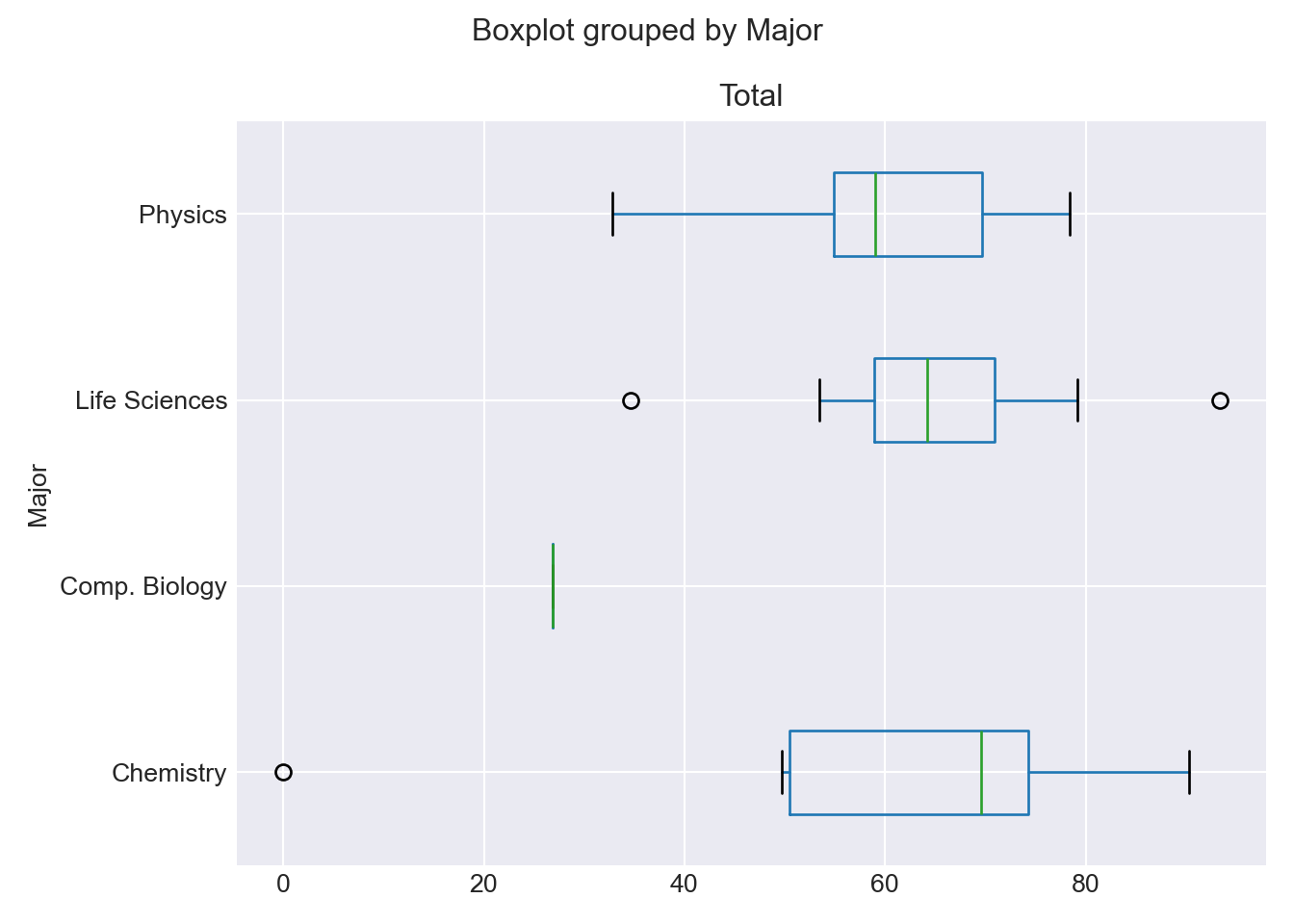
-
df_combined.boxplot(by='Major', column=['Test 1', 'Test 2', 'Test 3', 'Total'], vert=False, figsize=(8, 6)); plt.show()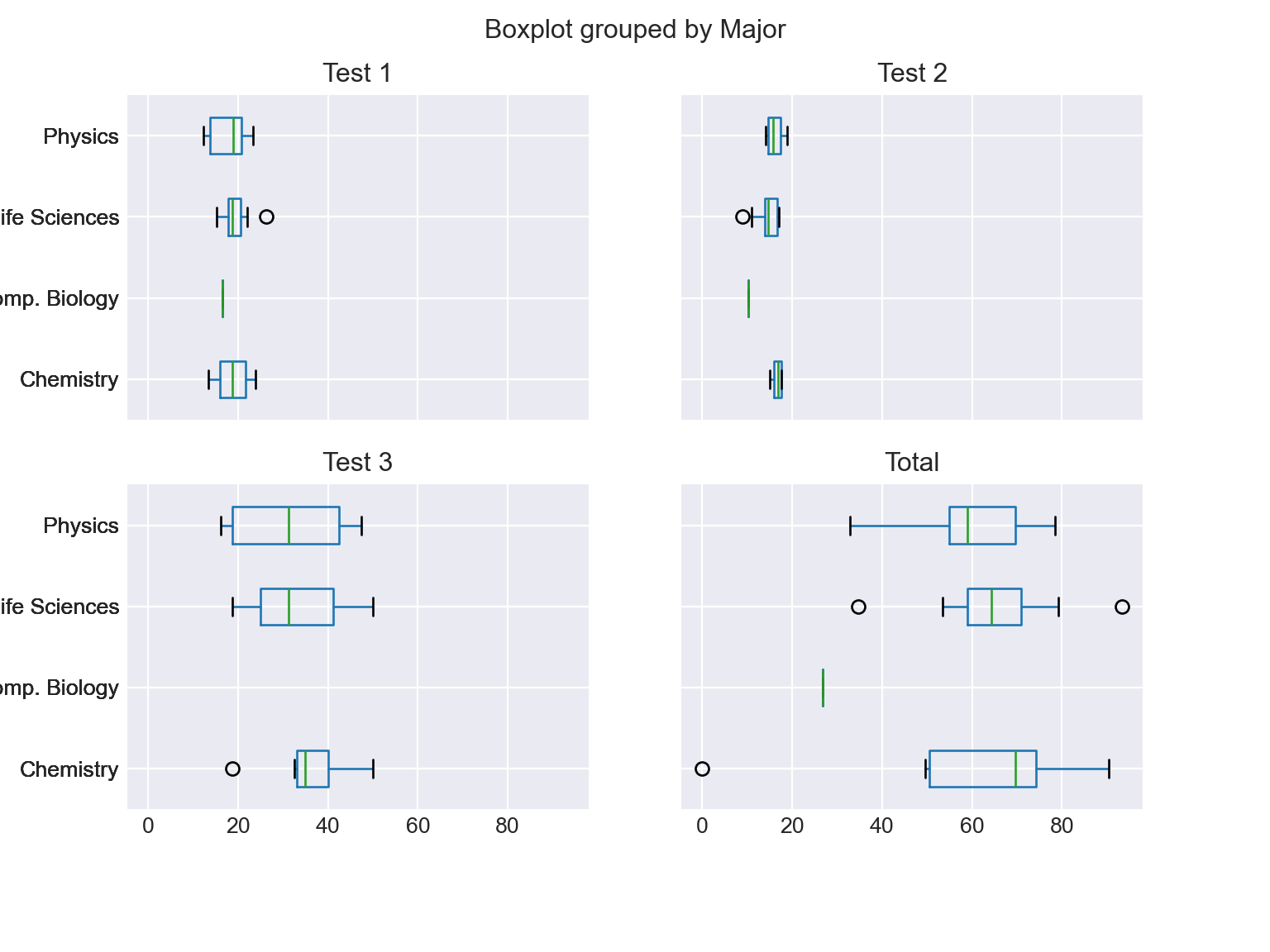
There is more that you can do; you can find more details about plotting with Pandas at visualisation page of the Pandas documentation.
